Fuzzy Connections - Introduction
The Fuzzy Connections and Multiple Sclerosis (MS) Lesion Finder tools have a great deal in
common. Both are designed to find fuzzy-connected features in images based
on the features' intensity characteristics. However, the Fuzzy
Connections tool is a general-purpose tool, while the MS Lesion Finder
has been tailored to the specific task of delineating MS lesions in MRI
scans of the head.
The algorithm used for the fuzzy connections is described in:
J.K.
Udupa and S. Samarasekera. "Fuzzy Connectedness and Object
Definition: Theory, Algorithms, and Application in Image
Segmentation". Graphical
Models and Image Processing
58:246-261 (1996).
Fuzzy connectivity is a seed-growing method: you define one or more regions on
interest that are all within the
feature you want to extract. The algorithm then grows the seed region(s)
based on the fuzzy affinity between pixels and their neighbours. The
fuzzy threshold setting then determines a cutoff fuzzy connectedness such
that all pixels above the threshold are considered part of the feature.
The affinity between pixels takes into account the degree of adjacency
between pixels as well as the similarity of their (multi-parametric)
intensity values. The result is a new set of ROIs that encompass the
fuzzy-connected feature.
Start the Fuzzy Connections and MS Lesion Finder tools from the
Toolkits menu:
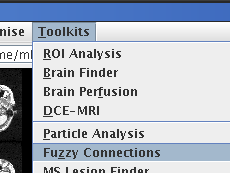
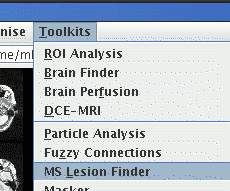
The Fuzzy Connections tool, shown below, is very similar in appearance to
the MS Lesion Finder.
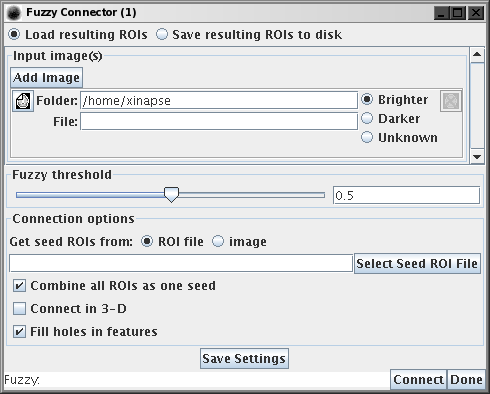
The setup which is common to both the Fuzzy Connections and MS Lesion
Finder tools is detailed below.
Input Images
Both tools can work one or more input images, where all images show the
same anatomical features in exactly the same pixel locations, but with
different image contrast. For example, these could be the two echoes
(proton-density-weighted and T2-weighted)
from a double-echo MRI sequence. Or they could be
T1-weighted and
T2-weighted images acquired sequentially but
with exactly the same data matrix and slice locations.
All input images must be of the same dimensionality (i.e., the same
number of slices, and rows and columns within the slice), and in order to
match the position of each pixel spatially, they must also have the same
pixel sizes and slice thicknesses. If you want to work with images that
have slightly different pixel positions or different pixel sizes, you may
be able to use the
Image Registration tool to bring all images into alignment and make
them have the same dimensionality. If your images are already aligned, but
have different numbers of pixel samples, then you can use the
Image Resampler tool to get the numbers
of samples to match across all images.
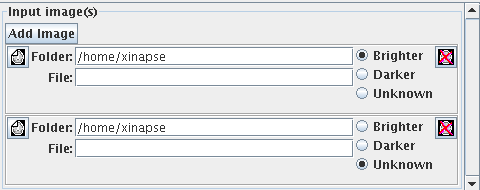
Click the  button until you
have the correct number of image selection panels. The figure below shows
the setup for working with two input images.
button until you
have the correct number of image selection panels. The figure below shows
the setup for working with two input images.
If you have too many images, you can remove any of them by clicking on the
 icon.
icon.
For each of the input images, set an input image file by
clicking on the load image icon:  . This will
bring up a File Chooser that you
can use to set the input image. Alternatively, you can simply type in the folder and
image file name in the text fields.
. This will
bring up a File Chooser that you
can use to set the input image. Alternatively, you can simply type in the folder and
image file name in the text fields.
For each input image, you can also provide an "intensity hint". To the
right of each input image, you will see:

You can use this to provide hints to the Fuzzy Connector about the
intensity characteristics of the fuzzy-connected feature you want to
extract. This can help to make the feature extraction more robust. Select:
 if the feature you want to extract is
always brighter than the surrounding background in this image.
if the feature you want to extract is
always brighter than the surrounding background in this image.
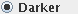 if the feature you want to extract is
always darker than the surrounding background in this image.
if the feature you want to extract is
always darker than the surrounding background in this image.
 if the feature you want to extract
varies in intensity relative to the surrounding background in this image, or if
you aren't sure.
if the feature you want to extract
varies in intensity relative to the surrounding background in this image, or if
you aren't sure.
Fuzzy Threshold
Next, set the fuzzy threshold. Selection of the right fuzzy threshold for
your particular problem is a matter of trial and error. Setting a lower
threshold results in a more generous inclusion of pixels in the extracted
feature (a larger feature), while setting a higher threshold results in a
more parsimonious inclusion (a smaller feature).

Set the fuzzy threshold by moving the slider, or by typing in a threshold
value. Fuzzy threshold values must lie between 0 an 1 (exclusive).
Seed pixels
Now tell the tool where to find the ROIs that are used to define the seed
pixels. The ROIs can either be read from a previously-saved ROI file, or
they can be read directly from the image displayed in Jim's main
display.
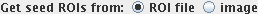
Note: if you are experimenting with different fuzzy thresholds
and other settings, it is strongly recommended that you save your seed
ROIs to a disk file, since the resulting feature is created as a set of
ROIs that can be automatically loaded onto the displayed image. Repeated
application of the Fuzzy Connector tool in the mode where seed ROIs are
read from the image will result in the
feature pixels being used as the seed pixels for the next fuzzy
connection application, which is presumably not what you would want.
Load one of your input images into
Jim's display.
Start the ROI Toolkit and define some ROIs
that are within the feature you want to delineate. Any
type(s) of ROI can be used. You can define a single ROI or multiple ROIs
as long as they are all contained within the feature. For ROIs that have
no area (Marker, Line and Curved Line ROIs), any pixels that the ROI
touches will be used as seed pixels. For other ROIs (Rectangular,
Elliptical or Irregular), pixels within the boundary of the ROI will be
used as seed pixels. Then either:
Connection Options
For information about setting the connection options, please see the
specific details for the Fuzzy
Connector tool or the MS Lesion
Finder tool.





 button until you
have the correct number of image selection panels. The figure below shows
the setup for working with two input images.
button until you
have the correct number of image selection panels. The figure below shows
the setup for working with two input images.

 icon.
icon.
 . This will
bring up a File Chooser that you
can use to set the input image. Alternatively, you can simply type in the folder and
image file name in the text fields.
. This will
bring up a File Chooser that you
can use to set the input image. Alternatively, you can simply type in the folder and
image file name in the text fields.

 if the feature you want to extract is
always brighter than the surrounding background in this image.
if the feature you want to extract is
always brighter than the surrounding background in this image.
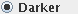 if the feature you want to extract is
always darker than the surrounding background in this image.
if the feature you want to extract is
always darker than the surrounding background in this image.
 if the feature you want to extract
varies in intensity relative to the surrounding background in this image, or if
you aren't sure.
if the feature you want to extract
varies in intensity relative to the surrounding background in this image, or if
you aren't sure.

 . Set the Fuzzy Connector to use
the ROI file you created by clicking the
. Set the Fuzzy Connector to use
the ROI file you created by clicking the  button and using the File Chooser to select the ROI file.
button and using the File Chooser to select the ROI file.
 . This will use the ROIs defined
on the displayed image to set the seed pixels.
. This will use the ROIs defined
on the displayed image to set the seed pixels.