Importing and Converting Images
Importing and Converting means changing a bunch of importable image files
(such as DICOM files) into one of the multi-slice image formats (such as
Analyze or UNC). The importing/converting process involves organising the
images so that the resulting Analyze or UNC images each represent one "scan".
What's the difference between importing and converting?
- Importing means loading a multi-slice image in an external format
(such as DICOM) directly into Jim's viewer. Therefore, only one
multi-slice image can be imported at a time. This is good for browsing
through images or analysing just one image at a time.
- Converting means changing a whole batch of multi-slice images in an external format
(such as DICOM) into a loadable format and
storing them on disk. This is good for handling images in bulk: they can
be conveniently loaded after conversion with minimal delay, and without
having re-import them for further analysis.
Jim can import/convert images in the following medical image file
formats:
- DICOM 3. Most DICOM 3 files can be imported. You may
experience difficulty if the images have been compressed in a
non-standard or unusual way. Please send mail to
 if you have difficulty.
if you have difficulty.
- ACR-NEMA I and II.
- General Electric (Genesis) format images. Please note that the
older GE 4.X format images are not currently supported.
- Siemens internal format images. These are the images that
usually have file names of the form XXXX-YY-ZZZ.ima.
- Bruker
ParaVision™ format images. A Bruker image data set is stored in
multiple files. Jim uses three of them to create an importable image:
- 2dseq contains the pixel data.
- reco contains details of how the image was reconstructed.
- method (higher up in the directory hierarchy) contains
details of image acquisition.
To correctly import Bruker images, Jim must have access to all 3 files.
You must therefore have the complete ParaVision™ directory tree structure
on your disk.
- Varian fdf (Flexible Data
Format) images.
If you have images in a format not currently supported, please contact
 to discuss your requirements.
to discuss your requirements.
To import/convert images in one of these formats, select Image Converter ...
from the File menu. 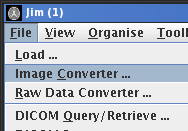 .
.
The Image Converter window will now appear:
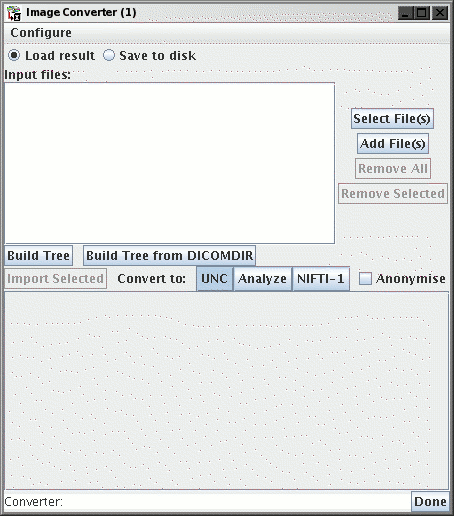
The Converter allows you to build a DICOM-like
Patient/Study/Series/Acquisition/Image Tree structure, from images
contained in a directory or set of directories. You can then
select a particular group of related images and load them as a
3-dimensional image data set.
Selecting the Images to be Converted
First, select the images files (such as DICOM files) to be
converted. Click the  button to choose the image files. You select either a directory
containing some files, or individual files within a directory (press the
SHIFT or CONTROL keys on your keyboard to select multiple directories or
files). To convert Bruker
ParaVision™ images, select the directory (or
directories) that contain the entire image data set(s). To convert
Varian images, select a either individual FDF files, or folder(s)
containing FDF files.
button to choose the image files. You select either a directory
containing some files, or individual files within a directory (press the
SHIFT or CONTROL keys on your keyboard to select multiple directories or
files). To convert Bruker
ParaVision™ images, select the directory (or
directories) that contain the entire image data set(s). To convert
Varian images, select a either individual FDF files, or folder(s)
containing FDF files.
You can add more files to the list by clicking the
 button.
Then, press the
button.
Then, press the  button.
button.
Alternatively, if you are reading your DICOM images from an offline
image source, such as a CD-ROM, there may be a DICOMDIR file on the
disk. A DICOMDIR file lists all the scans that are present on the
CD-ROM, and scanning the DICOMDIR file is much quicker than reading all
the image header. Click the
 button to
use this method of building the image tree. A File Chooser will pop up, allowing you to
select the DICOMDIR on the CD-ROM. The tree build will then begin.
button to
use this method of building the image tree. A File Chooser will pop up, allowing you to
select the DICOMDIR on the CD-ROM. The tree build will then begin.
After scanning through the files you have chosen, the Converter will
build a "tree" structure of patients and scans. Double click
on one of the patients listed to show the individual image series
(scans) for that patient, as shown below for patient "V.C".
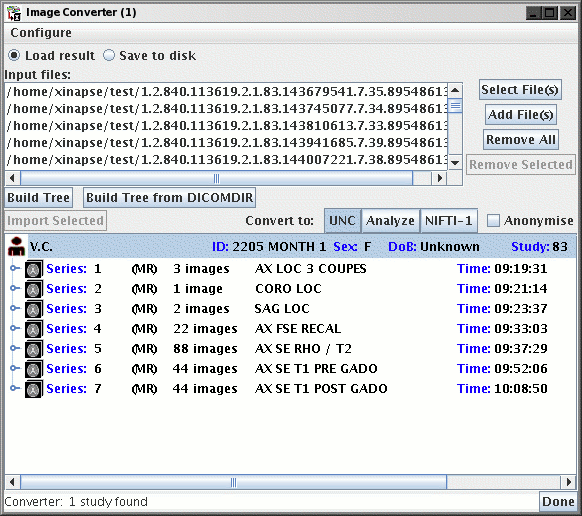
Double clicking on any part of the list either expands of collapses that
part of the list.
Importing or Converting
To import images (load into the Jim's viewer), select "Load
result". To batch convert images, select "Save to disk".

- To import an image, ensure you have a node of the image tree containing only
one 3-dimensional image dataset (such as one image series), as shown
below. Then click the
 button.
button.
If the  is not
active, this means that you have not selected a tree node that contains
only one 3-D dataset. Double click on that tree node to expand it, and
select one of the sub-nodes of the tree. After a few moments, the
multi-slice image that you selected will now be loaded into the Image
Display frame, and the image source text box, near the top of the Image
Display frame will show a suggested file name for the loaded image.
is not
active, this means that you have not selected a tree node that contains
only one 3-D dataset. Double click on that tree node to expand it, and
select one of the sub-nodes of the tree. After a few moments, the
multi-slice image that you selected will now be loaded into the Image
Display frame, and the image source text box, near the top of the Image
Display frame will show a suggested file name for the loaded image.
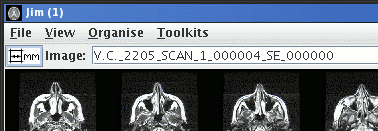
This is the name that will be suggested for the image should you
decide to save the image in one of the loadable formats. The suggested file name is
composed from a combination of the patient name, patient ID, study
number, series number and acquisition number for the scan.
Note that the imported data is not saved to disk in a loadable
format unless you explicitly do so.
- To convert images, select any node of the image tree.
All images below that node will be converted. For example, if you select a
patient node, all scans for that patient will be converted to a 3-D
format and stored on disk. You can select more than one node by pressing
the SHIFT or CONTROL keys as you click on the tree nodes.
one 3-dimensional image dataset (such as one image series), as shown
below.
Choose the format of the images that will be
created, by clicking on the appropriate button:

Then click the  button to start the conversion. The images will be stored in Jim's
current working directory. To change the image storage directory select
button to start the conversion. The images will be stored in Jim's
current working directory. To change the image storage directory select
Image Storage Directory from the Configure menu
(see below).
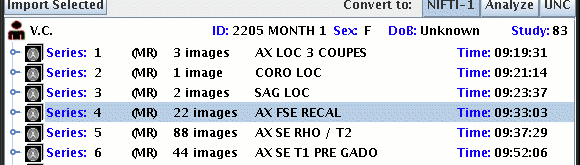
If you do not want to import or convert all slices of a particular scan,
then double click on the series node for that scan to expand it and show
the nodes for the individual image slices. If a series has multiple
acquisitions, then double click on the acquisition node also. Use the
SHIFT and CONTROL keys on your keyboard as you click on the slices you
want (to select more than one slice). In the example below, the CONTROL
key was press as the mouse was clicked on the individual slices, to select
just slices 1, 3, 5 and 6 of Series 4.
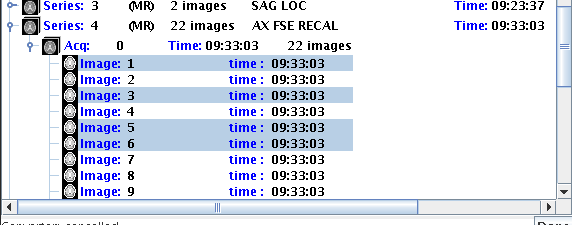
Press the SHIFT key as you click to select a complete range of slices.
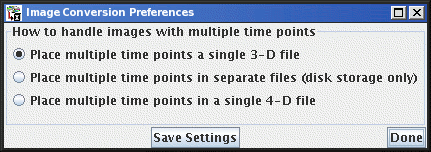
Anonymising the Converted Images
If you want to anonymise the patient details, select the
 check box before
you start the conversion or import. The patient name and other patient
details (such as the patient's maiden name, address and 'phone number)
will be stripped from the images as they are created.
In addition, the file name of the image created will not contain the
patient's name.
check box before
you start the conversion or import. The patient name and other patient
details (such as the patient's maiden name, address and 'phone number)
will be stripped from the images as they are created.
In addition, the file name of the image created will not contain the
patient's name.
Configuring Image Converter
The following options are available under the Configure menu:
Click the  button when you have
finished with importing or converting images.
button when you have
finished with importing or converting images.
 if you have difficulty.
if you have difficulty.
 if you have difficulty.
if you have difficulty.
 to discuss your requirements.
to discuss your requirements.
 .
.

 button to choose the image files. You select either a directory
containing some files, or individual files within a directory (press the
SHIFT or CONTROL keys on your keyboard to select multiple directories or
files). To convert Bruker
ParaVision™ images, select the directory (or
directories) that contain the entire image data set(s). To convert
Varian images, select a either individual FDF files, or folder(s)
containing FDF files.
button to choose the image files. You select either a directory
containing some files, or individual files within a directory (press the
SHIFT or CONTROL keys on your keyboard to select multiple directories or
files). To convert Bruker
ParaVision™ images, select the directory (or
directories) that contain the entire image data set(s). To convert
Varian images, select a either individual FDF files, or folder(s)
containing FDF files.
 button.
Then, press the
button.
Then, press the  button.
button.
 button to
use this method of building the image tree. A File Chooser will pop up, allowing you to
select the DICOMDIR on the CD-ROM. The tree build will then begin.
button to
use this method of building the image tree. A File Chooser will pop up, allowing you to
select the DICOMDIR on the CD-ROM. The tree build will then begin.


 button.
button.

 is not
active, this means that you have not selected a tree node that contains
only one 3-D dataset. Double click on that tree node to expand it, and
select one of the sub-nodes of the tree. After a few moments, the
multi-slice image that you selected will now be loaded into the Image
Display frame, and the image source text box, near the top of the Image
Display frame will show a suggested file name for the loaded image.
is not
active, this means that you have not selected a tree node that contains
only one 3-D dataset. Double click on that tree node to expand it, and
select one of the sub-nodes of the tree. After a few moments, the
multi-slice image that you selected will now be loaded into the Image
Display frame, and the image source text box, near the top of the Image
Display frame will show a suggested file name for the loaded image.


 button to start the conversion. The images will be stored in Jim's
current working directory. To change the image storage directory select
button to start the conversion. The images will be stored in Jim's
current working directory. To change the image storage directory select

 check box before
you start the conversion or import. The patient name and other patient
details (such as the patient's maiden name, address and 'phone number)
will be stripped from the images as they are created.
In addition, the file name of the image created will not contain the
patient's name.
check box before
you start the conversion or import. The patient name and other patient
details (such as the patient's maiden name, address and 'phone number)
will be stripped from the images as they are created.
In addition, the file name of the image created will not contain the
patient's name.

 button when you have
finished with importing or converting images.
button when you have
finished with importing or converting images.