Brain Perfusion Analysis
Having set up your input and output images as described in the Introduction, you can now set up the
quantification parameters for brain perfusion. In the Perfusion
Analysis tool, you will see:
Quantification parameters
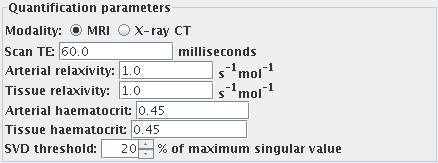
First, set the imaging modality by selecting one of:
 if you are working with
Magnetic Resonance Imaging (MRI) data.
if you are working with
Magnetic Resonance Imaging (MRI) data.
 if you are working with
X-ray Computed Tomography (X-ray CT) data.
if you are working with
X-ray Computed Tomography (X-ray CT) data.
This determines which quantification parameters should be set.
It is not necessary to set the quantification
parameters if you want semi-quantitative analysis to be
performed. Semi-quantitative analysis will allow you to obtain
relative perfusion parameters, not absolute cerebral blood flow
(CBF) and cerebral blood volume (CBV). However, the mean transit
time (MTT) is truly quantitative regardless of the settings of
these parameters.
N.B. it is very difficult to obtain the values you need to
enter for true quantification. These values depend very much on
your pulse sequence and contrast agent, and Xinapse Systems does
not, in general, recommend that you try to obtain quantitative
perfusion parameters.
Now set the following parameters, if applicable:
- Use brain finder. If you select this option, the Brain Finder tool will attempt to
isolate the brain before perfusion analysis is performed. For this to
work, the dataset you are analysing must be a multi-slice axial set
covering much of the brain. This option is not available for X-ray CT
image analysis - it will be greyed out.
- Scan TE (MRI data only). The scan is assumed to be T2 or
T2*-weighted. For quantification, enter the correct
echo time.
- Arterial relaxivity (relaxivity of the contrast agent
for large vessels) (MRI data only). This is the slope of the plot of
R2* versus concentration of contrast agent in
the blood plasma in the large vessels. Needed
for quantification, otherwise leave as 1.0.
- Tissue relaxivity (relaxivity of the contrast agent for
the capillary bed) (MRI data only). This is the slope of the plot of
R2* versus concentration of contrast agent in
the blood plasma in the capillaries. Needed
for quantification, otherwise leave as 1.0.
- Arterial haematocrit. The haematocrit in large vessels. Needed
for quantification, otherwise leave as 0.45.
- Tissue haematocrit. The haematocrit in the capillary bed. Needed
for quantification, otherwise leave as 0.45.
- SVD threshold. This affects the sensitivity of the analysis to
noise and pseudo-random variation in the input images. Increasing
this value will make the analysis more immune to noise, at the
expense of bias in the quantification. The default of 40% should
be fine for most images.
Arterial Input Function (AIF)
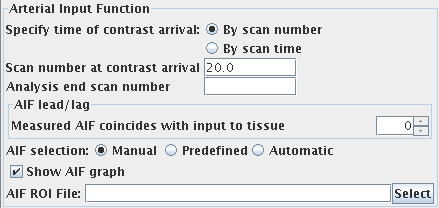
To obtain an arterial input function (the concentration of
contrast agent in an artery that feeds the brain), you must set
the following:
Brain Perfusion Output Parametric Images
The Brain Perfusion analysis produces four output images, that
have the same base name, but have the output parameter name
appended. These are:
- CBF. Cerebral blood flow, with units of mls blood / 100g tissue / minute.
- CBV. Cerebral blood volume, with units that indicate the blood
volume as a percentage of the total tissue volume.
- MTT. The mean transit time, in seconds.
- RSq. A measure of the goodness of fit. This is the sum of the
squares of the residuals between the fitted curve and the
time-series data.
These output images are in floating-point format, and can be
viewed using Jim. The output images will be of the same
image type as the first input image.
Example output images from the Brain Perfusion Tool.
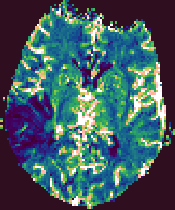 |
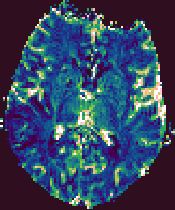 |
 |
| Cerebral Blood Flow |
Cerebral Blood Volume |
Mean Transit Time |


 if you are working with
Magnetic Resonance Imaging (MRI) data.
if you are working with
Magnetic Resonance Imaging (MRI) data.
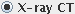 if you are working with
X-ray Computed Tomography (X-ray CT) data.
if you are working with
X-ray Computed Tomography (X-ray CT) data.

 , you can
specify this either as:
, you can
specify this either as:
 , you can
specify this either as:
, you can
specify this either as:
 button, or type the name into
the text field:
button, or type the name into
the text field:

 button, or type the name into
the text field:
button, or type the name into
the text field:

![]()