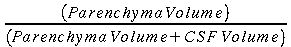Brain Finder - Calculating the BPF
Having outlined the brain in
every image slice, you can now compute the brain parenchymal
fraction, an index of brain atrophy. The computation involves
selection of an intensity threshold that best separates the brain
parenchyma from the CSF surrounding the brain and in the
ventricles. This is done by examining the pixel intensity
histogram, and separating the parenchyma peak from the CSF peak.
Note: if you have previously created the brain outline, and
saved the ROIs to disk, you can simply
reload them from disk at
this stage, without re-finding them.
When performing fully-automated analysis, the BPF calculation has two
parameters that may need to be adjusted, depending on the type of image
being analysed.
Note: fully-automated analysis is strongly recommended to aid
consistency of BPF measurements.
Fitting Separate Grey- and White-Matter Peaks
Some, more heavily T1-weighted, images have two peaks for the
parenchyma: one for the grey matter, and one for the white matter. If this
is the case for your images, then select

To check whether this setting is appropriate for your images, you
need to view the image intensity histogram of the brain. First mask the image to isolate just the brain
and CSF. Then view the intensity histogram for the whole image by opening the
image statistics dialog. Below is
a 3-D FLASH image that requires separate peaks fitted to the grey
and white matter.

| 
|
| Heavily T1-weighted 3-D FLASH image.
| Intensity histogram for the image to the left (all
slices), with separate peaks for CSF, grey matter and white matter.
|
Setting the Threshold Between CSF and Parenchyma
When calculating the BPF, an intensity threshold is set which divides CSF
from brain: all pixel with intensity below the threshold are considered to
be CSF, and all those with intensity above are considered to be brain. The
threshold is located between the histogram peaks for the CSF and brain (or
grey-matter, if separately fitting GM and WM). The exact threshold
location is determined by the CSF/brain threshold fraction:
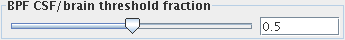
Use the slider to set a value between 0 and 1. A value of 0 will put the
threshold at the position of the CSF peak in the intensity histogram.
A value of 1 will put the threshold at the position of the brain (or GM) peak in the
intensity histogram. A setting of 0.5 (the default value) will put
threshold half way between the CSF and brain peaks.
If you find that too much brain tissue is being classed as CSF (the BPF is
too low), then reduce the threshold fraction. Conversely,
if you find that too much CSF is being classed as brain tissue (the BPF is
too high), then increase the threshold fraction.
Note: whatever setting you use your images, for both
cross-sectional and serial studies it is important to maintain the
same setting for all patients scanned on any given
scanner. Because of differences in contrast from scanner to
scanner, it may be necessary to adjust the settings on a
scanner-by-scanner basis, but for serial studies it is imperative
that the same setting is always used for any given patient/scanner.
You also have three further options:
- If you do not want the CSF / parenchyma intensity threshold
to be selected automatically, and you have another method for
determining the threshold, then select:

and enter the threshold in the field provided. Any pixels with intensities above
the threshold will be counted as parenchyma, and those below will be
counted as CSF.
Note: when the threshold is set manually, no further image
uniformity correction is performed as part of the BPF computation.
- If it is clear from the result that BPF calculation is problematic,
then you can help Jim to find the BPF by manually setting the
initial guesses for the CSF, and parenchyma peak positions. Click the "Set
initial guesses for peak locations manually" check box.

Using the whole-brain intensity histogram as a guide, enter your estimates
of the peak positions for the CSF and parenchyma or (if you are fitting
grey and white matter peaks separately) grey and white matter peaks.
- For the purposes of verification, you can create images
showing the distribution of parenchyma and CSF. To create these
images, select
 .
Images are created with the original image name plus suffixes "CSF" and "Brain"
(or, if you are fitting grey and white matter peaks separately) "GM" and "WM".
.
Images are created with the original image name plus suffixes "CSF" and "Brain"
(or, if you are fitting grey and white matter peaks separately) "GM" and "WM".
Finally, click on the  button. This
fits a distribution of intensities to the histogram, assigns the
peaks to CSF and parenchyma (or CSF, grey matter and white matter,
if selected) and then chooses the intensity threshold between the
CSF and parenchyma (or grey matter) peak positions according to the BPF
CSF/brain threshold fraction (see above).
The threshold is then applied to separate CSF from parenchyma, and
the BPF calculated as:
button. This
fits a distribution of intensities to the histogram, assigns the
peaks to CSF and parenchyma (or CSF, grey matter and white matter,
if selected) and then chooses the intensity threshold between the
CSF and parenchyma (or grey matter) peak positions according to the BPF
CSF/brain threshold fraction (see above).
The threshold is then applied to separate CSF from parenchyma, and
the BPF calculated as:
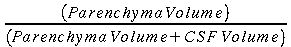
When complete, a window pops up showing the result, and giving you
the opportunity to write these results as a permanent record to
disk in a report.
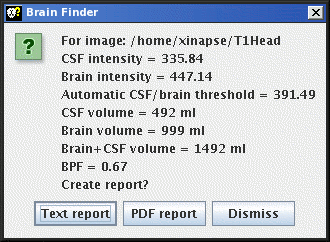
- If you choose to write to a text file report, then a File chooser will appear, for you
to choose a log file name. The default file extension for log
files is ".log". If the chosen file already exists, a BPF
report entry will be appended to the log file.
The format of a Brain Finder log file is given in the
file formats section.
- If you choose to write to a
PDF file report, then a File chooser will appear, for you
to choose a PDF file name. If the chosen file already exists, a Brain Finder
report entry will be appended to the PDF file.
- Select the "Dismiss" option if you do not want to write a report.
A graph will also pop up showing the quality of the fit that is used to
estimate the CSF/brain threshold.
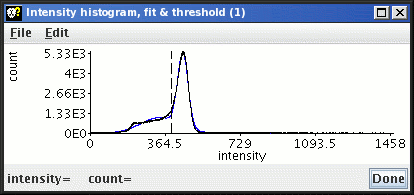
The black line represents the image intensity histogram (after
uniformity correction); the blue line represents the fit to the two (or
three) peaks in the histogram; and the vertical dashed line is the
threshold that divides CSF from brain.






 .
Images are created with the original image name plus suffixes "CSF" and "Brain"
(or, if you are fitting grey and white matter peaks separately) "GM" and "WM".
.
Images are created with the original image name plus suffixes "CSF" and "Brain"
(or, if you are fitting grey and white matter peaks separately) "GM" and "WM".
 button. This
fits a distribution of intensities to the histogram, assigns the
peaks to CSF and parenchyma (or CSF, grey matter and white matter,
if selected) and then chooses the intensity threshold between the
CSF and parenchyma (or grey matter) peak positions according to the BPF
CSF/brain threshold fraction (see above).
The threshold is then applied to separate CSF from parenchyma, and
the BPF calculated as:
button. This
fits a distribution of intensities to the histogram, assigns the
peaks to CSF and parenchyma (or CSF, grey matter and white matter,
if selected) and then chooses the intensity threshold between the
CSF and parenchyma (or grey matter) peak positions according to the BPF
CSF/brain threshold fraction (see above).
The threshold is then applied to separate CSF from parenchyma, and
the BPF calculated as: